Gene Ontology primer
Gene ontology (geneontology.org) is a controlled vocabulary of terms used for annotation of genes. Terms are connected into hierarchy showing their relationships. For GO enrichment analysis in particular the most important term relationships are is_a and part_of. You may consider terms connected with this type of relationship as being 'successor/predecessor' of one another. Here is a small snapshot of ontology tree.
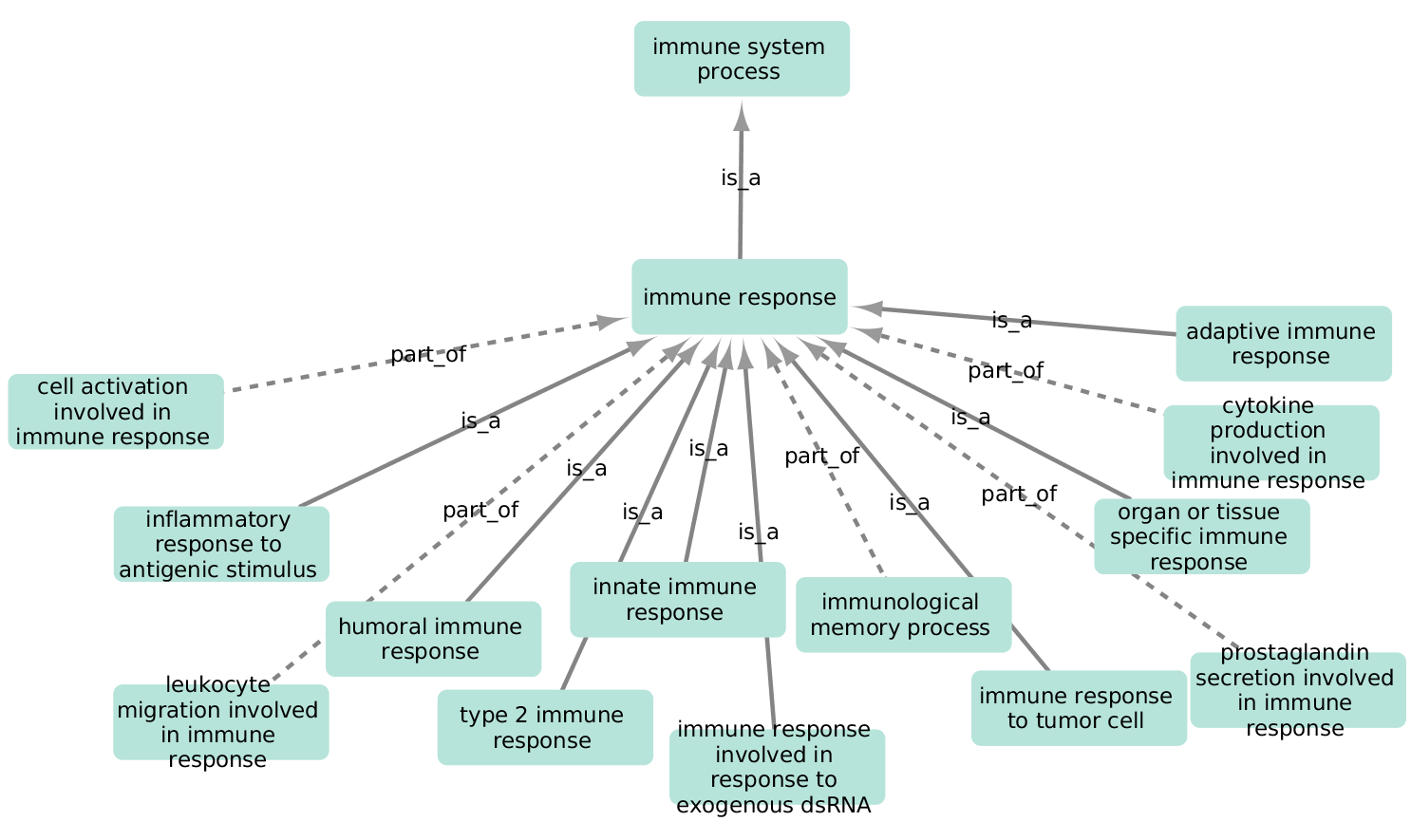
Here GO term "immune response" (GO:0006955) has 13 child GO terms: "cell activation involved in immune response" (GO:0002263), "inflammatory response to antigenic stimulus" (GO:0002437) etc. Important to note that this hierarchy implies the possibility to propagate annotation for a specific gene up the tree to the root ("immune system process" in this case). This means that a gene involved in e.g. "innate immune response" is also annotated by all of its ancestors i.e. "immune response" and "immune system process".
GOnet analysis
The application is capable of constructing an interactive graph containing GO terms and genes. The graph will simultaneously convey hierarchical structure of the GO terms and connect GO terms to genes according to available annotations.